Reproducible analysis pipelines are a crucial part of the day-to-day operation of the Bioinformatics Core; ensuring that we can process data for researchers in an efficient manner and have an auditable trail of what tools (and versions) have been used.
Nextflow is a popular modern workflow management tool that SBC has been adopting recently. A workflow management tool takes care of the interaction with the High-Performance Computing (HPC) system to ensure that new analyses are run as required, and can even cope without resuming from failed runs. This makes running the workflow as hands-off as possible. Nextflow also uses containerisation technologies (such as docker or apptainer) to ease the process of software installation.
Whilst other workflow managers are available (e.g. snakemake), for us the main attraction of using nextflow is that many common Bioinformatics pipelines have been implemented using nextflow and are available via the community-curated nf-core.re website. Below is a schematic diagram of the current RNA-seq pipeline
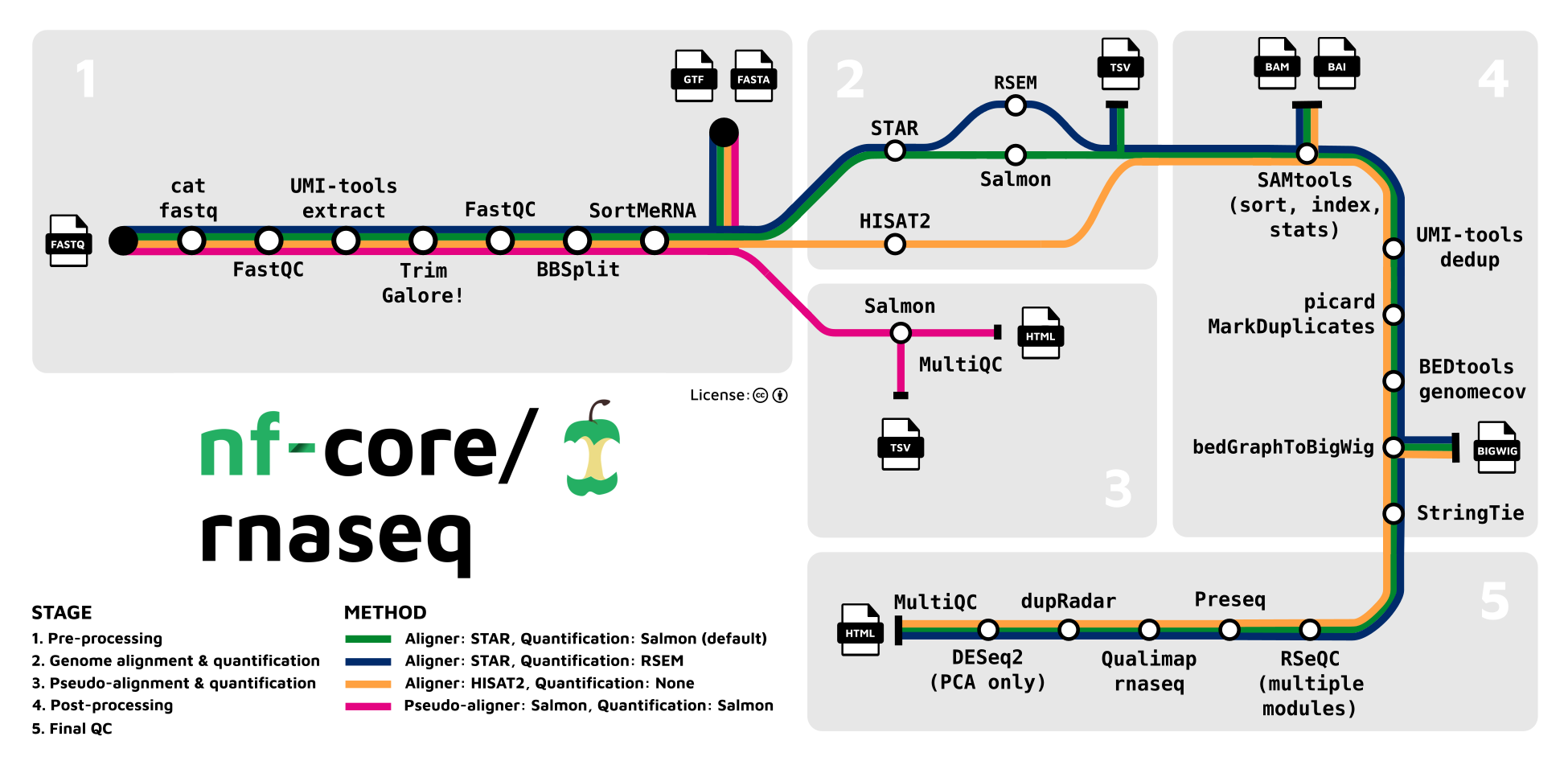
Having these pipelines available to run “out of the box” means that we are able to deliver results to researchers quicker without having to develop and maintain our own pipelines.
Moreover, researchers wishing to run their own NGS analysis can benefit from nextflow. Developing analysis pipelines is not something that we would recommend for most researchers when a best-practice solution is already available. We are liaising with Research Software Engineering and Research-IT teams at University of Sheffield to make nextflow pipelines available to all users of HPC. When this work is completed, researchers should be able to create a simple samplesheet describing their biological samples, and be able to analyse these samples using a gold-standard pipeline with minimal prior knowledge of command-line Bioinformatics.
In due course, we will make documentation available on running nf-core workflows on the University of Sheffield HPC, but please get in touch if you would like more information.
For queries relating to collaborating with the Bioinformatics Core team on projects: bioinformatics-core@sheffield.ac.uk
Join our mailing list so as to be notified when we advertise talks and workshops by subscribing to this Google Group. You can also connect with us on Linkedin.
Requests for a Bioinformatics support clinic can be made via the Research Software Engineering (RSE) code clinic system. This is monitored by Bioinformatics Core staff, so we will ensure the appropriate expertise (which may involve individuals from multiple teams) will be available to help you
Queries regarding sequencing and library preparation provision at The University of Sheffield should be directed to the Multi-omics facility in SITraN or the Genomics Laboratory in Biosciences.
