The Sheffield Bioinformatics Core offers in- depth bioinformatic analysis of single cell RNA sequencing datasets. This exciting area of analysis allows us to identify and measure heterogeneity within populations of cells and further mine subpopulations for differentially expressed cell marker genes and enriched pathways.
Our custom analysis can include:
We have recently contributed our single cell analysis to two exciting publications. The first characterised the zebrafish endogenous retrovirus, zferv, and hypothesised about its role in immunity and brain development [1]. In this study we were able to integrate previously published datasets to compare expression of zferv1a across haematopoietic lineages.
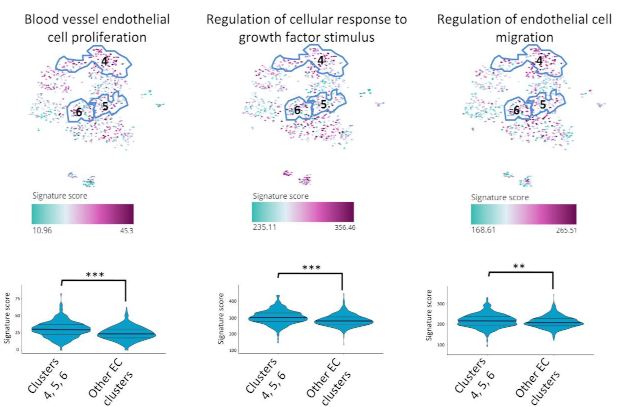
In the second, the JAG1-NOTCH4 pathway was examined in its role in atherosclerosis [2]. Sub- clustering of single cell rna-seq data from murine heart cells were able to show that JAG1 expression suppresses endothelial subsets with roles in vascular repair.
As the field of single cell transcriptomics progresses, demand for high quality analysis and collaboration increases. Please get in touch with the SBC to chat about your single cell analysis - especially if you are preparing a grant application that will require Bioinformatics support.
We are also planning some training workshops on the analysis of single-cell (10X) data. Please see our training page for updates.
[1] Rutherford HA, Clarke A, Chambers EV, Petts JJ, Carson EG, Isles HM, Duque-Jaramillo A, Renshaw SA, Levraud JP, Hamilton N. A zebrafish reporter line reveals immune and neuronal expression of endogenous retrovirus. Dis Model Mech. 2022 Apr 1;15(4):dmm048921. doi: 10.1242/dmm.048921. Epub 2022 Apr 13. PMID: 35142349; PMCID: PMC9016899.
[2] Souilhol C, Tardajos Ayllon B, Li X, Diagbouga MR, Zhou Z, Canham L, Roddie H, Pirri D, Chambers EV, Dunning MJ, Ariaans M, Li J, Fang Y, Jørgensen HF, Simons M, Krams R, Waltenberger J, Fragiadaki M, Ridger V, De Val S, Francis SE, Chico TJ, Serbanovic-Canic J, Evans PC. JAG1-NOTCH4 mechanosensing drives atherosclerosis. Sci Adv. 2022 Sep 2;8(35):eabo7958. doi: 10.1126/sciadv.abo7958. Epub 2022 Aug 31. PMID: 36044575; PMCID: PMC9432841.
For queries relating to collaborating with the Bioinformatics Core team on projects: bioinformatics-core@sheffield.ac.uk
Join our mailing list so as to be notified when we advertise talks and workshops by subscribing to this Google Group. You can also connect with us on Linkedin.
Requests for a Bioinformatics support clinic can be made via the Research Software Engineering (RSE) code clinic system. This is monitored by Bioinformatics Core staff, so we will ensure the appropriate expertise (which may involve individuals from multiple teams) will be available to help you
Queries regarding sequencing and library preparation provision at The University of Sheffield should be directed to the Multi-omics facility in SITraN or the Genomics Laboratory in Biosciences.
